alternative promoters
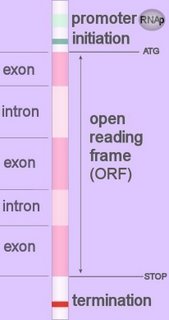 Promoters direct specialized enzymes to the location at which to commence reading the segment of DNA that codes for production of a protein.
Promoters direct specialized enzymes to the location at which to commence reading the segment of DNA that codes for production of a protein.Promoters can be divided into two broad categories: those without and those with CpG islands — stretches of DNA containing multiple copies of the nonmethlated dinucleotide CpG, which consists of the nucleotide cytosine (C) followed by guanine (G).
CpG islands comprise about 1 to 2% of the mammalian genome, and about 56% of sequenced human genes have CpG islands near their 5′ ends. This includes all genes that are ubiquitously expressed (housekeeping genes) plus many genes with a tissue-restricted pattern of expression. Promoters are normally located at the upstream edge of the CpG island, such that one or more of the 5′ exons of the gene generally fall within the island region. Although most CpG islands are nonmethylated in all tissues, a small proportion of islands become methylated during development [s].
In adults, single promoters with CpG islands tend to be linked to “housekeeping” genes, whereas single promoters without CpG islands are more often associated with highly regulated biological systems such as the immune and digestive systems.
A greater than expected percentage of mammalian genes have alternative promoters, which are more active during embryological development. Roughly 40-50 % of human and mouse genes have alternative promoters.
Alternative promoters can produce the same protein as single promoters, yet can be active in different tissues or at different times. In other cases, alternative promoters direct the polymerase enzymes to commence reading DNA at different start codons, ultimately resulting in different proteins with different functions.
Alternative promoters can affect gene expression in diverse ways. Production of different mRNA isoforms may be effected directly through different transcription start sites or indirectly through promoter-directed exon inclusion. The resulting transcripts may encode different protein isoforms, or they may vary only in their 5’ untranslated regions, affecting mRNA stability and translation efficiency.
Some genes employ promoters that differ in strength to direct tissue specific expression. Because tight regulation is essential for accurate gene function, loss of regulatory control can have serious disease-causing phenotypic effects such as malignancies resulting from activation of alternative promoters.
Alternative promoters, which confer greater flexibility, are more stable than single promoters over evolutionary time. Mammalian genomes are highly conserved and it is widely believed that gene regulation is largely responsible for the diversity of form and function between species. The higher evolutionary conservation of alternative promoters reflects the higher density of functional elements involved in regulating promoter choice.
Alternative promoters are tightly regulated, in line with their importance in cellular function. Cells with more than one promoter regulate which promoter to use, and when. Alternative promoters are more active during embryonic development.
Alternative Ways of Reading DNA Have Spurred Evolution : Green's studies of promoter function suggest intriguing hypotheses about evolutionary patterns. “The way C. elegans and a lot of other organisms increase their flexibility is simply to make a duplicate copy of a gene and have different regulation for the duplicate,” said Green. “With mammals, one way in which evolution has generated more diversity is to produce different versions of the same gene and allow the cell to regulate expression in multiple tissues. This is a way that evolution gets more bang for its buck, because it gets additional functions for the same gene.”
Labels: alternative promoter, conservation, CpG island, embryologic development, evolution, housekeeping genes, RNA polymerase










































<< Home